Research
Our Research
INSEPTION group focuses on hallmarks of tumor evolution including:
- the increased cellular proliferative capacity,
- the invasive mechanisms that cancer cells adopt, varying from single cell migration to collective strategies,
- the metabolic reprogramming,
- the extensive genetic and phenotypic heterogeneity, and
- the formation of new vasculature and remodeling of the extracellular matrix (ECM).
Tumors are not growing in isolation. The tumor microenvironment plays a significant role in regulating dynamically the invasive and proliferative capacities of cells. Both irregular vascularization and matrix stiffness have been observed in tumor microenvironment to affect tumor progression, invasion, drug delivery, drug resistance and metastasis. On the other hand, the metabolic alterations of cancer cells have also a dramatic effect on the extracellular microenvironment, generating a dynamic feedback between the cells and their microenvironment. The investigation of cell-to-cell, cell-to-matrix and cell-drug interactions at several levels and scales of complexity, comprise the main focus of exploration in our work.
Highlights
- Subcellular, metabolic footprints determine the fate of tumor evolution (https://www.doi.org/10.1109/JBHI.2018.2890708)
- Cohesive, invasive patterns seen for the first time in primary GB spheroids (https://www.doi.org/10.1038/s41598-018-34521-5)
- In vivo Molecular Imaging approaches monitor tumor evolution: perspectives and limitations (https://www.doi.org/10.1109/IST.2014.6958483)
- Integrating in vitro & in silico approaches to evaluate anti-cancer drug therapy (https://www.doi.org/10.1109/EMBC.2016.7592130)
- Develop an in silico glioma model to explore the competition between proliferative and invasive cancer phenotypes (https://www.doi.org/10.1371/journal.pone.0103191)
Our approach towards preclinically driven models
Our multidisciplinary approach involves the use of different Glioblastoma models to identify the basic mechanisms and describe their contribution in Glioblastoma progress by using computational modeling. More specifically, it combines the use of patient-specific computer-based models together with both in vitro and in vivo brain cancer models, including patient-derived models and clinical studies. Tissue from naïve patients with high grade brain cancer is excised, as routinely done. Part of this tissue is used for the orthotopic/ heterotopic transplantation of immunodeficient mice. Samples from the same tissue are also used for 3D cell cultures generation. These cultures are pharmaceutically screened with chemotherapeutic agents and also scanned to image physiologic biomarkers with advanced imaging modalities. All data collected are translated and integrated to initialize, parametrize and validate glioma growth computational algorithms, leading to pre-clinically driven models. Such an approach enables tight control of experimental parameters and high reproducibility. We can eventually verify the precise set of their computational counterparts that are needed towards a systematic in silico mapping of cancer progression.
New cancer markers and methods, as they develop by current genomic technologies, may be incorporated in our models to foster personalized care with less medical costs.
Research directions
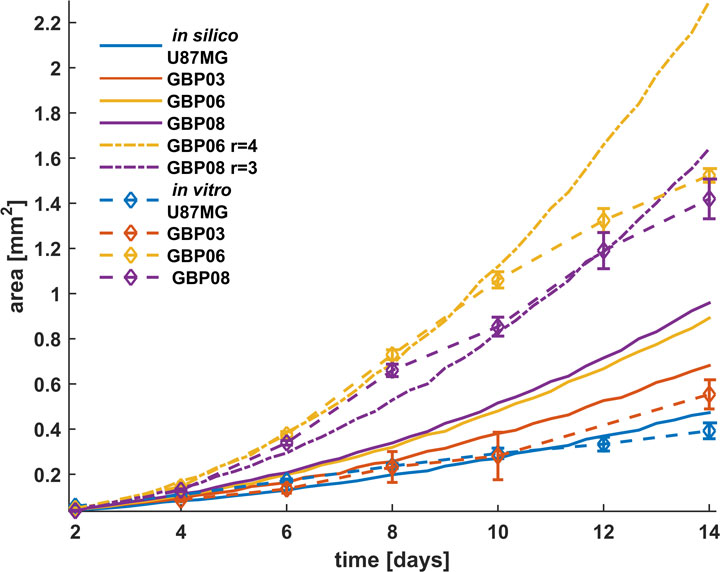
Tumor growth over time
Excessive proliferation is the most crucial cancer hallmark independently to the cancer type. Glioblastoma (GB) tumors have a remarkable rapid growth that has a critical role regarding the space-occupation and the development of intracranial pressure, usually the main reason of the GB symptomatology. As it is well-understood, both cell division and local spreading are responsible for cancer expansion comprising the most important aspects for cancer progress.
We utilize both primary and secondary GB cells that are cultivated in vitro as 3D tumor spheroids and computational approaches to study, experimentally parametrize and predict the growth dynamics of tumor spheroids focusing on proliferation. The importance of quantitative methods to provide spatial information of proliferative, quiescent and necrotic cells, as well as additional features including the remodeling of the extracellular matrix and phenotypic distribution regarding intra-tumoral heterogeneity affecting tumor expansion becomes evident.
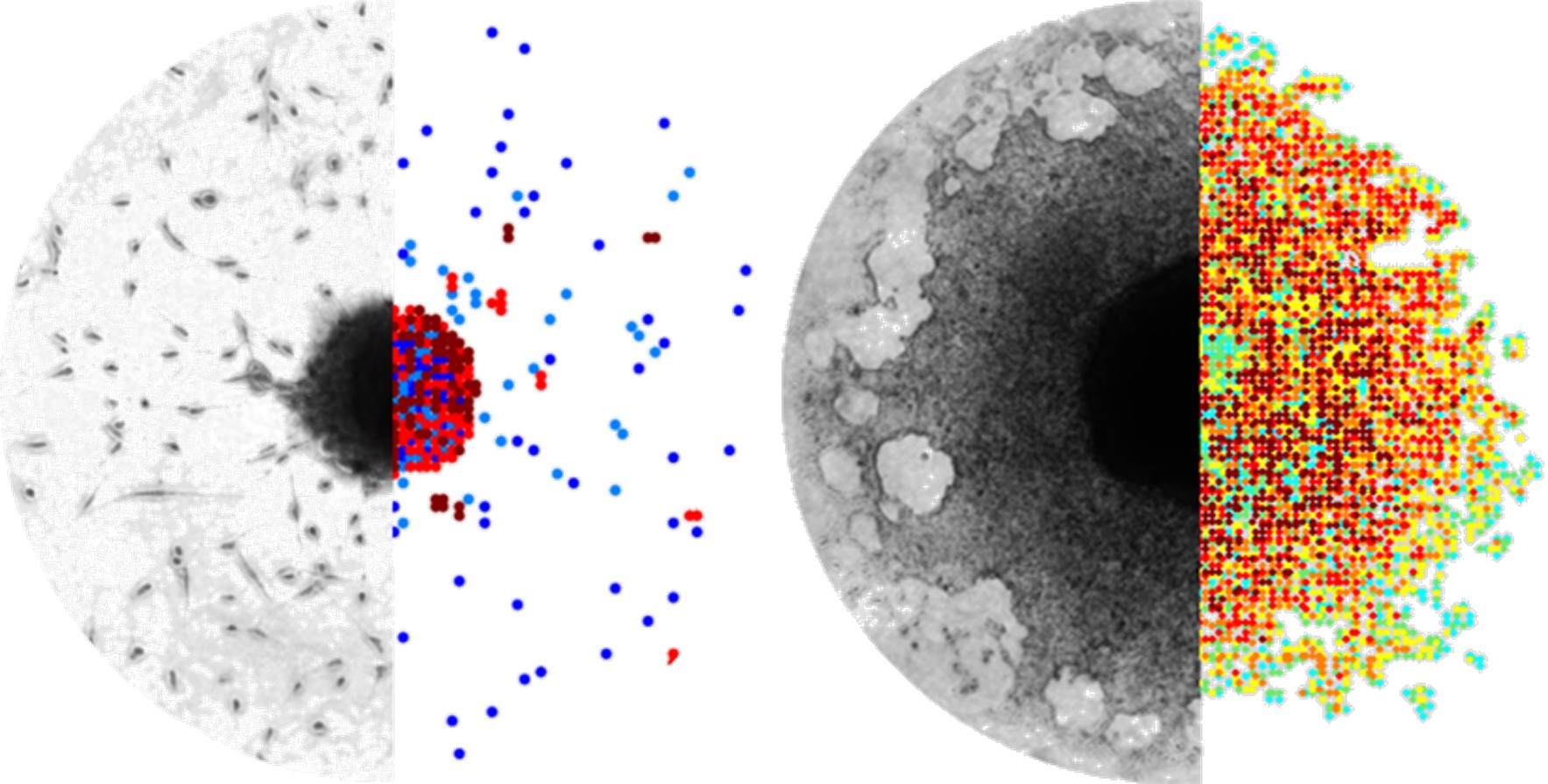
Tumor invasion
Invasion is a complex, multiscale phenomenon involving processes at different spatial and temporal scales. Migrating tumor cells can mechanistically move by different modes, ranging from single cell to collective locomotion, or even to whole-tissue expansion. The molecular pathways during movement are complex and involve both energy utilization and response to stimuli, either chemical or mechanical or both. The invasive process necessitates both locomotion and proteolysis and involves both cell-to-matrix and cell-to-cell adhesion mechanisms.
We explore the possible underlying mechanisms involved in the distinct invasive patterns adopted by different primary and secondary Glioblastoma cell lines. Our mathematical approaches question the potential role of the intrinsic heterogeneity with respect to cell-to-cell/ matrix adhesion on tumor morphology and growth dynamics.
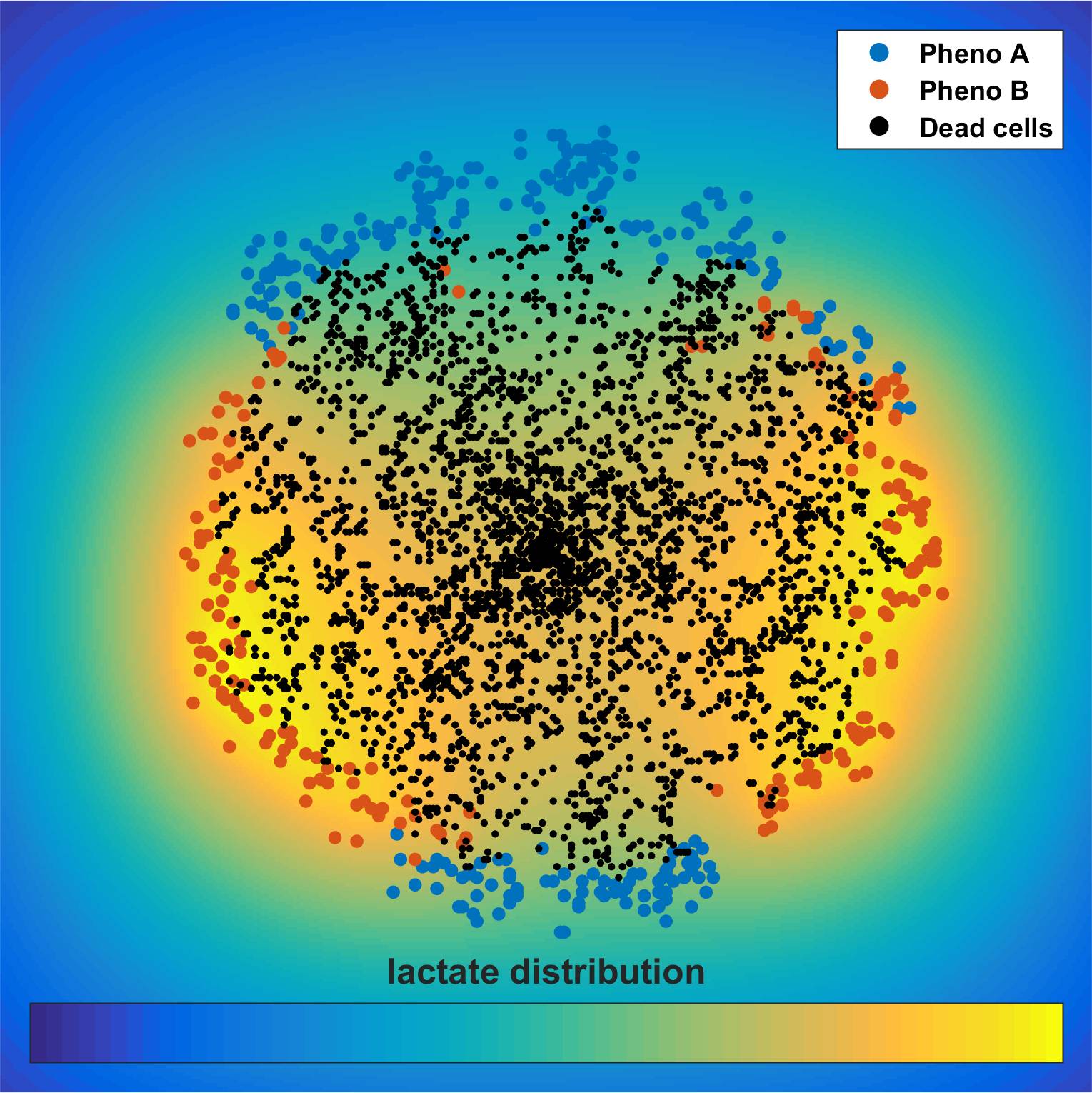
Cancer metabolism
In contrast to normal cells, cancer cells produce a substantial amount of energy inefficiently, metabolizing glucose to lactate independent of oxygen availability, a phenomenon termed as Warburg effect or aerobic glycolysis. Although the aberrant metabolism of cancer cells was initially observed almost a century ago, metabolic reprogramming has been only recently included in the hallmarks of cancer. The exact regulatory mechanisms of cancer metabolism are far from complete. The tumor microenvironment alters metabolite transporters and glycolytic enzymes, while signaling pathways, involving a number of oncogenes and tumor suppressor genes have been found to be implicated in the altered metabolism. The metabolic state of a cell affects not only its own decision-making, but the metabolic alterations of cancer cells have also a dramatic effect on the extracellular microenvironment, generating a dynamic feedback between the cells and their microenvironment. In order to study the effects of sub-cellular characteristics on tumor evolution, we incorporate sub-cellular information utilizing existing genome-scale metabolic modeling approaches that link genotypes with phenotypes into cell-centered tumor growth models that account for the spatiotemporally heterogeneous tumor microenvironment and show that genotypes with different metabolic footprint can have dramatically different tumor expansion patterns.
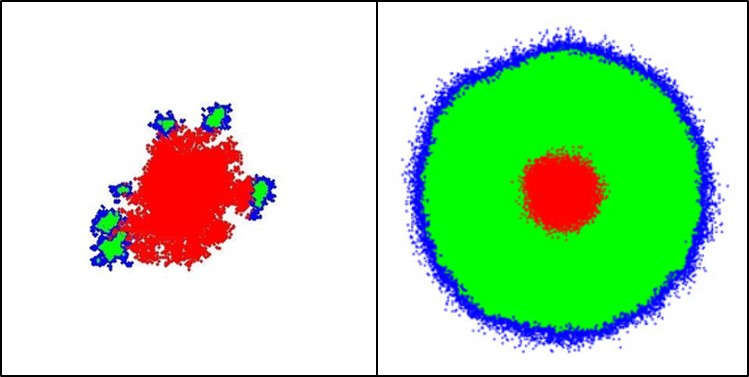
The role of cell death in Glioblastoma progress
Interestingly, during the observation of the MR images of the GB patients, as well as by taking into account the empirical clinical knowledge, the next remark is noted: the vast majority of the Glioblastoma (GB) tumor within the brain lesion is necrotic. However, this necrotic pattern is common between all GB cases and can be attributed in both hypoxia formation due to nutrient-deficiency, as well as to another critical feature of the tumor growth, the intrinsic cell death probability of the cancer cells. This spontaneous GB cell death is considered to be mainly necrosis, rather than apoptosis or autophagy.
Assessing the pathophysiology of GB necrosis formation and evolution, as well as fostering the intrinsic glioma cell death is a current treatment target to several studies and will further promote our GB ontogenesis understanding and the importance of necrosis as a GB diagnostic feature.
Preclinical drug screening
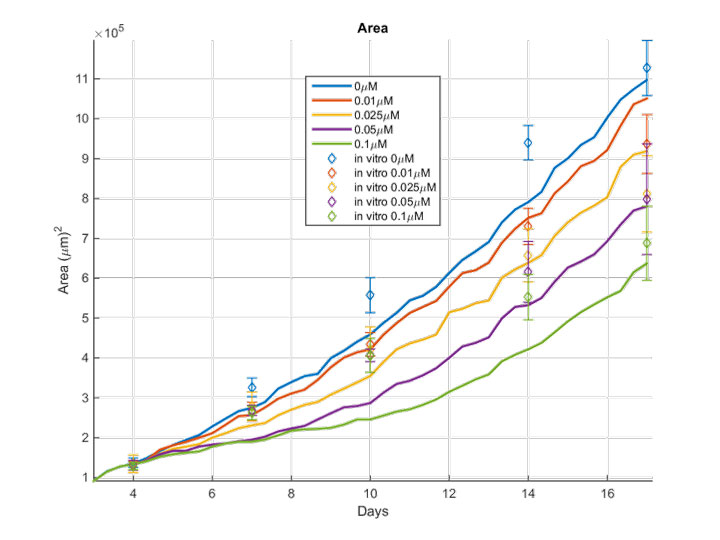
Glioblastoma (GB) prognosis remains poor mainly because of the high inter- and intra-tumoral heterogeneity and post-surgery relapse. GB recent trends in preclinical drug screening usually employ first advanced in vitro models utilizing patient-derived GB cells. Biologically-motivated in silico methods are proposed to expand our insight regarding the pharmacodynamics of several drug agents on primary GB cells and predict the spatiotemporal response to therapy. Given the growth inhibiting effects observed in vitro for the drug agents tested, as well as the reported mechanisms of action, we computationally investigate whether our biological results can be further discriminated and extrapolated to a combined drug therapeutic scheme. This way we enable a drug screening tool that would otherwise be time-consuming and costly to be experimentally tested, while would raise ethical concerns to be clinically applied.
The role of PML in Glioblastoma evolution
The Promyelocytic Leukemia Protein (PML) is a cell regulator, expressed in all tissues and modifications in its expression are often related to various carcinogenic phenotypes. However, the specific effects of this protein in cancer are not yet clear because it can have either tumor suppressing or tumor promoting effects depending on the type of cancer. In brain, PML participates in the physiological migration of the neural progenitor cells, which are also considered as the cell of origin of Glioblastoma (GB). Furthermore, it has been hypothesized that in GB, PML affects the evolution of the tumor by inhibiting the growth rate of the cells, while also inducing cellular migration.
We study the role of PML in GB physiology using 3D in vitro and ex vivo biological models along with 3D optical imaging. Our findings are integrated with cancer predictive algorithms, in order to study further the PML-mediated mechanisms that underlie our observations.